vector movement and Py5vector#
This tutorial is best paired with the tutorial on Object-Oriented Programming, and will use the code created within that tutorial. If you aren’t yet familiar with this concept, it’s worth going back and reading through the OOP tutorial.
The word vector is used in a few different ways, so let’s clarify: these are not the vectors used in vector graphics, but Euclidean vectors, also known as geometric vectors or spatial vectors. Essentially, a vector is just a way of measuring direction and magnitude (or strength) of movement.
When we’re discussing movement in a real or virtual space, we might refer to the force at which something moves, or the speed, or the distance. If we give something (say, an amoeba, as we’ll be using in the following exercises) ten seconds to move, the distance that it covers in that time will depend on how fast it’s going (which depends on the strength of the force that was pushing it that way). This strength of movement is the magnitude of the vector. It doesn’t inherently say anything about the distance traveled, or the direction, but it can tell you how hard something is being “pushed” in that direction.
If the amoeba below is going to move 400 pixels to the right, we don’t necessarily know how fast it’s moving. However, we do know its direction – to the right, along the X axis of this two-dimensional space. If we wanted to move it this direction in py5, we’d say that its X position is increasing.
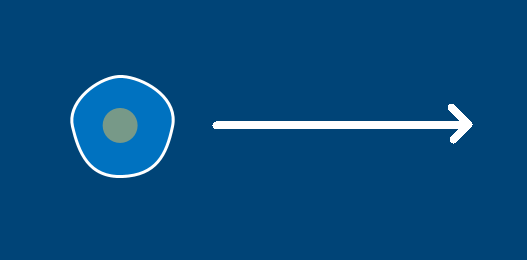
Of course, much of the time directional movement isn’t along just a single axis. The amoeba below is moving up and to the right, so it’s moving along both the X and Y axis. Again, even without knowing its magnitude, we can at least know its direction. In py5, its X position would be increasing and its Y position would be decreasing. To accomodate for this sort of diagonal movement, two-dimensional vectors can store both an X and a Y value. In this case, the vector might be (1, -1) to represent movement to the right and upwards. You might not know its magnitude just by looking at it, but you can use the .mag method (appended to the end of any vector) to find out. In general, it’s enough to point your vector in the right direction, and increase the values (like (3, -3)) to increase the magnitude of movement.
We’ll be using the amoeba code from our OOP tutorial to start us off. Depending on if you want to have your Amoeba class as a separate module file, you’ll either have two files (my suggestions for names are at the top of the following code snippets) or a single, consolidated, all-in-one file. Either will work.
# amoeba.py
from py5 import *
cs = get_current_sketch()
class Amoeba():
def __init__(self, x, y, diameter):
self.x = x
self.y = y
self.d = diameter
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.x + self.nucleus['x'],
self.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(cs.frame_count/(r/2), r/8)
xp, xm = self.x+cpx, self.x-cpx
yp, ym = self.y+cpy, self.y-cpy
begin_shape()
vertex(
self.x, self.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.x+r, self.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.x, self.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.x-r, self.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.x, self.y-r # (back to) top vertex
)
end_shape()
# microscope.py
from amoeba import Amoeba
bob = Amoeba(400, 200, 100)
def setup():
size(800, 400)
def draw():
background('#004477')
bob.display()
# all-in-one file
class Amoeba():
def __init__(self, x, y, diameter):
self.x = x
self.y = y
self.d = diameter
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.x + self.nucleus['x'],
self.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(frame_count/(r/2), r/8)
xp, xm = self.x+cpx, self.x-cpx
yp, ym = self.y+cpy, self.y-cpy
begin_shape()
vertex(
self.x, self.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.x+r, self.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.x, self.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.x-r, self.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.x, self.y-r # (back to) top vertex
)
end_shape()
bob = Amoeba(400, 200, 100)
def setup():
size(800, 400)
def draw():
background('#004477')
bob.display()
Whatever method you use, the above code will display a single amoeba, Bob, on the screen. Let’s define a new vector and use it to give Bob some movement.
First, we’ll be adding two new parameters, xspeed and yspeed, to our Amoeba class’s initialization method. We’ll then use these to form a new attribute, propulsion, that determines our amoeba’s movement.
We’ll also need to pass those new arguments in when Bob is created. Next, in our draw() function in the main sketch, we’ll actually adjust Bob’s X and Y position based on that propulsion stat.
# all-in-one file
class Amoeba():
def __init__(self, x, y, diameter, xspeed, yspeed):
self.x = x
self.y = y
self.d = diameter
self.propulsion = Py5Vector(xspeed, yspeed) # Look! A vector!
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.x + self.nucleus['x'],
self.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(frame_count/(r/2), r/8)
xp, xm = self.x+cpx, self.x-cpx
yp, ym = self.y+cpy, self.y-cpy
begin_shape()
vertex(
self.x, self.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.x+r, self.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.x, self.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.x-r, self.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.x, self.y-r # (back to) top vertex
)
end_shape()
bob = Amoeba(400, 200, 100, 3, -1)
def setup():
size(800, 400)
def draw():
background('#004477')
bob.x += bob.propulsion.x
bob.y += bob.propulsion.y
bob.display()
Run this sketch, and Bob will fly off the screen. Each frame, their X position is increasing by 3 pixels, and their Y position is decreasing by 1 pixel.
We’ve got a lot of code referencing Bob’s X and Y position now, and you can notice that we add to these values separately on these lines:
bob.x += bob.propulsion.x
bob.y += bob.propulsion.y
If we also store the amoeba’s position with a vector, we can do this a little more efficiently. First, let’s define a new attribute, location, inside of __init__(), and use it instead of those x and y variables. After that, we’ll just have to find and replace each instance of self.x and self.y with self.location.x and self.location.y. (If you’re using a dedicated IDE or even a text editing program, you may have a find-and-replace feature to make things easier.)
After that, those two lines can be replaced with just one:
bob.location += bob.propulsion
# all-in-one file
class Amoeba():
def __init__(self, x, y, diameter, xspeed, yspeed):
self.location = Py5Vector(x, y)
self.d = diameter
self.propulsion = Py5Vector(xspeed, yspeed) # Look! A vector!
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.location.x + self.nucleus['x'],
self.location.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(frame_count/(r/2), r/8)
xp, xm = self.location.x+cpx, self.location.x-cpx
yp, ym = self.location.y+cpy, self.location.y-cpy
begin_shape()
vertex(
self.location.x, self.location.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.location.x+r, self.location.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.location.x, self.location.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.location.x-r, self.location.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.location.x, self.location.y-r # (back to) top vertex
)
end_shape()
bob = Amoeba(400, 200, 100, 3, -1)
def setup():
size(800, 400)
def draw():
background('#004477')
bob.location += bob.propulsion
bob.display()
If you like, you can add additional forces to this simulation by adding more vectors. For example, since many single-celled organisms live in water, you could define some vector for a current and add this current to Bob’s location each frame, too. Of course, in addition to adding vectors, you can always subtract them. This can be useful for the reasons you might expect (like a force that pushes your amoeba in the opposite direction), but we can also use it to force our amoebas to move towards the mouse pointer.
In py5, we have access to the mouse_x and mouse_y variables, which track the position of the mouse cursor. Since we’re using vectors, and thus directions (instead of absolute positions), we won’t simply be moving the amoeba to this location; we want to move it towards the position of mouse_x and mouse_y. This means that we’ll do a bit of subtraction to find the difference in space between the current location of the amoeba and the position of the mouse, and move the amoeba in that direction.
# all-in-one file
class Amoeba():
def __init__(self, x, y, diameter, xspeed, yspeed):
self.location = Py5Vector(x, y)
self.d = diameter
self.propulsion = Py5Vector(xspeed, yspeed) # Look! A vector!
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.location.x + self.nucleus['x'],
self.location.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(frame_count/(r/2), r/8)
xp, xm = self.location.x+cpx, self.location.x-cpx
yp, ym = self.location.y+cpy, self.location.y-cpy
begin_shape()
vertex(
self.location.x, self.location.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.location.x+r, self.location.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.location.x, self.location.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.location.x-r, self.location.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.location.x, self.location.y-r # (back to) top vertex
)
end_shape()
bob = Amoeba(400, 200, 100, 3, -1)
def setup():
size(800, 400)
def draw():
background('#004477')
pointer = Py5Vector(mouse_x, mouse_y)
difference = pointer - bob.location
bob.location += difference
bob.display()
Currently, the magnitude of this movement is a little too high. Bob will instantly “jump” to the mouse pointer, and stay there.

Luckily, there’s a method for this. We can use the .set_limit() method on any vector to limit its magnitude. By setting the limit of our difference vector to a small number, we can make this a bit more sensible.
# all-in-one file
class Amoeba():
def __init__(self, x, y, diameter, xspeed, yspeed):
self.location = Py5Vector(x, y)
self.d = diameter
self.propulsion = Py5Vector(xspeed, yspeed) # Look! A vector!
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.location.x + self.nucleus['x'],
self.location.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(frame_count/(r/2), r/8)
xp, xm = self.location.x+cpx, self.location.x-cpx
yp, ym = self.location.y+cpy, self.location.y-cpy
begin_shape()
vertex(
self.location.x, self.location.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.location.x+r, self.location.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.location.x, self.location.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.location.x-r, self.location.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.location.x, self.location.y-r # (back to) top vertex
)
end_shape()
bob = Amoeba(400, 200, 100, 3, -1)
def setup():
size(800, 400)
def draw():
background('#004477')
pointer = Py5Vector(mouse_x, mouse_y)
difference = pointer - bob.location
bob.location += difference.set_limit(1)
bob.display()
Now, the movement added to the amoeba’s location each frame is clamped at a sensibly low value.

We have one amoeba on the screen, but there’s no reason we can’t have a dozen. You could manually go through naming each amoeba, creating it, updating its position… and so on, and so forth. However, we can automate this process using a for loop. We’ll replace our current line to create Bob with a loop creating many amoebas (and storing them in a list), and then replace our lines in draw() to move Bob with a loop accessing this list of amoebas and calling the correct methods on all of them.
# all-in-one file
class Amoeba():
def __init__(self, x, y, diameter, xspeed, yspeed):
self.location = Py5Vector(x, y)
self.d = diameter
self.propulsion = Py5Vector(xspeed, yspeed) # Look! A vector!
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.location.x + self.nucleus['x'],
self.location.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(frame_count/(r/2), r/8)
xp, xm = self.location.x+cpx, self.location.x-cpx
yp, ym = self.location.y+cpy, self.location.y-cpy
begin_shape()
vertex(
self.location.x, self.location.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.location.x+r, self.location.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.location.x, self.location.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.location.x-r, self.location.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.location.x, self.location.y-r # (back to) top vertex
)
end_shape()
amoebas = []
for i in range(12):
diameter = random(50, 200)
speed = 1000 / (diameter * 50)
x, y = random(800), random(400)
amoebas.append(Amoeba(x, y, diameter, speed, speed))
def setup():
size(800, 400)
def draw():
background('#004477')
pointer = Py5Vector(mouse_x, mouse_y)
for a in amoebas:
difference = pointer - a.location
a.location += difference.set_limit(1)
a.display()
So that we can fully appreciate the differing movements of all of these randomized amoebas, we’ll make a couple of changes. First, let’s give each amoeba another attribute, a direction vector (created with the arguments xdirection and ydirection), that will kick in when it isn’t following the mouse.
So that we actually get to see this behavior, we’ll also add a new if statement where the difference is being calculated. If the mouse is being held down, the amoebas will move towards it. Otherwise, they can use this new direction vector to move towards instead.
# all-in-one file
class Amoeba():
def __init__(self, x, y, diameter, xspeed, yspeed, xdirection, ydirection):
self.direction = Py5Vector(xdirection, ydirection)
self.location = Py5Vector(x, y)
self.d = diameter
self.propulsion = Py5Vector(xspeed, yspeed) # Look! A vector!
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.location.x + self.nucleus['x'],
self.location.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(frame_count/(r/2), r/8)
xp, xm = self.location.x+cpx, self.location.x-cpx
yp, ym = self.location.y+cpy, self.location.y-cpy
begin_shape()
vertex(
self.location.x, self.location.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.location.x+r, self.location.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.location.x, self.location.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.location.x-r, self.location.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.location.x, self.location.y-r # (back to) top vertex
)
end_shape()
amoebas = []
for i in range(12):
diameter = random(50, 200)
speed = 1000 / (diameter * 50)
x, y = random(800), random(400)
xdirection, ydirection = random(-5, 5), random(-5, 5)
amoebas.append(Amoeba(x, y, diameter, speed, speed, xdirection, ydirection))
def setup():
size(800, 400)
def draw():
background('#004477')
pointer = Py5Vector(mouse_x, mouse_y)
for a in amoebas:
if is_mouse_pressed:
difference = pointer - a.location
else:
difference = a.direction
a.location += difference.set_limit(1)
a.display()
Of course, now the amoebas are liable to drift off the screen entirely, and it can take a while for them to move back to your mouse cursor. To fix this, we can add some code so that if an amoeba exits the screen, it reappears on the opposite side of the window, preserving its original momentum.
# all-in-one file
class Amoeba():
def __init__(self, x, y, diameter, xspeed, yspeed, xdirection, ydirection):
self.direction = Py5Vector(xdirection, ydirection)
self.location = Py5Vector(x, y)
self.d = diameter
self.propulsion = Py5Vector(xspeed, yspeed) # Look! A vector!
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.location.x + self.nucleus['x'],
self.location.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(frame_count/(r/2), r/8)
xp, xm = self.location.x+cpx, self.location.x-cpx
yp, ym = self.location.y+cpy, self.location.y-cpy
begin_shape()
vertex(
self.location.x, self.location.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.location.x+r, self.location.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.location.x, self.location.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.location.x-r, self.location.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.location.x, self.location.y-r # (back to) top vertex
)
end_shape()
amoebas = []
for i in range(12):
diameter = random(50, 200)
speed = 1000 / (diameter * 50)
x, y = random(800), random(400)
xdirection, ydirection = random(-5, 5), random(-5, 5)
amoebas.append(Amoeba(x, y, diameter, speed, speed, xdirection, ydirection))
def setup():
size(800, 400)
def draw():
background('#004477')
pointer = Py5Vector(mouse_x, mouse_y)
for a in amoebas:
if is_mouse_pressed:
difference = pointer - a.location
else:
difference = a.direction
a.location += difference.set_limit(1)
# Wrapping around to the opposite edge of the screen!
r = a.d / 2
if a.location.x - r > width:
a.location.x = 0 - r
if a.location.x + r < 0:
a.location.x = width + r
if a.location.y - r > height:
a.location.y = 0 - r
if a.location.y + r < 0:
a.location.y = height + r
a.display()
These new if statements check the X and Y positions of the amoeba, and its radius, against the size of the window (with the 0 values representing the top and left sides of the screen, and height and width representing the bottom and right sides). If an amoeba crosses over, they’ll appear on the opposite side of the screen – but the position they’re heading towards is preserved, since it relies on the direction of a vector!
collision detection task#
Currently, the amoebas can overlap freely, but it would be interesting if they avoided or even bounced off of each other. Your challenge is to add a collision detection feature to facilitate this.
To test whether an amoeba is overlapping another amoeba, you’ll add another for loop inside of the a in amoebas loop. (Remember, a is a name we decide ourselves, to refer to a single amoeba in the loop.) We’ll also want to add some code to prevent trying to compare an amoeba’s location to itself. This will use the continue keyword, which moves to the next iteration of the loop.
# all-in-one file
class Amoeba():
def __init__(self, x, y, diameter, xspeed, yspeed, xdirection, ydirection):
self.direction = Py5Vector(xdirection, ydirection)
self.location = Py5Vector(x, y)
self.d = diameter
self.propulsion = Py5Vector(xspeed, yspeed) # Look! A vector!
self.nucleus = {
'fill': ['#FF0000', '#FF9900', '#FFFF00',
'#00FF00', '#0099FF'][int(random(5))],
'x': self.d * random(-0.15, 0.15),
'y': self.d * random(-0.15, 0.15),
'd': self.d / random(2.5, 4)
}
def circle_point(self, t, r):
x = cos(t) * r
y = sin(t) * r
return [x, y]
def display(self):
# nucleus
fill(self.nucleus['fill'])
no_stroke()
circle(
self.location.x + self.nucleus['x'],
self.location.y + self.nucleus['y'],
self.nucleus['d'])
# cell membrane
fill(0x880099FF)
stroke('#FFFFFF')
stroke_weight(3)
r = self.d / 2.0
cpl = r * 0.55
cpx, cpy = self.circle_point(frame_count/(r/2), r/8)
xp, xm = self.location.x+cpx, self.location.x-cpx
yp, ym = self.location.y+cpy, self.location.y-cpy
begin_shape()
vertex(
self.location.x, self.location.y-r # top vertex
)
bezier_vertex(
xp+cpl, yp-r, xm+r, ym-cpl,
self.location.x+r, self.location.y # right vertex
)
bezier_vertex(
xp+r, yp+cpl, xm+cpl, ym+r,
self.location.x, self.location.y+r # bottom vertex
)
bezier_vertex(
xp-cpl, yp+r, xm-r, ym+cpl,
self.location.x-r, self.location.y # left vertex
)
bezier_vertex(
xp-r, yp-cpl, xm-cpl, ym-r,
self.location.x, self.location.y-r # (back to) top vertex
)
end_shape()
amoebas = []
for i in range(10):
diameter = random(50, 200)
speed = 1000 / (diameter * 50)
x, y = random(800), random(400)
xdirection, ydirection = random(-5, 5), random(-5, 5)
amoebas.append(Amoeba(x, y, diameter, speed, speed, xdirection, ydirection))
def setup():
size(800, 400)
def draw():
background('#004477')
pointer = Py5Vector(mouse_x, mouse_y)
for a in amoebas:
if is_mouse_pressed:
difference = pointer - a.location
else:
difference = a.direction
a.location += difference.set_limit(1)
# Wrapping around to the opposite edge of the screen!
r = a.d / 2
if a.location.x - r > width:
a.location.x = 0 - r
if a.location.x + r < 0:
a.location.x = width + r
if a.location.y - r > height:
a.location.y = 0 - r
if a.location.y + r < 0:
a.location.y = height + r
a.display()
for b in amoebas:
if a is b:
continue
# ... otherwise, what should happen?
Think about how you might use a vector to push apart two overlapping amoeba. You can use the radius of each amoeba (which you might already know you can get by dividing the diameter of each amoeba by two - a.d/2 and b.d/2) to determine if they’re overlapping. If the distance between their locations is less than their radii added together, they’re overlapping. Otherwise, they’re far enough away to avoid an overlap.
To test what happens when an overlap does occur, you can always draw just two amoebas (by changing the loop to use range(2) and get them to move towards your mouse cursor until they intersect.
You may find it helpful to draw a line between their locations, or to calculate a distance vector somehow and print the value of distance.mag to see how it changes as they move closer to each other.
Good luck!